The Biology of CRISPR-Cas Immunity
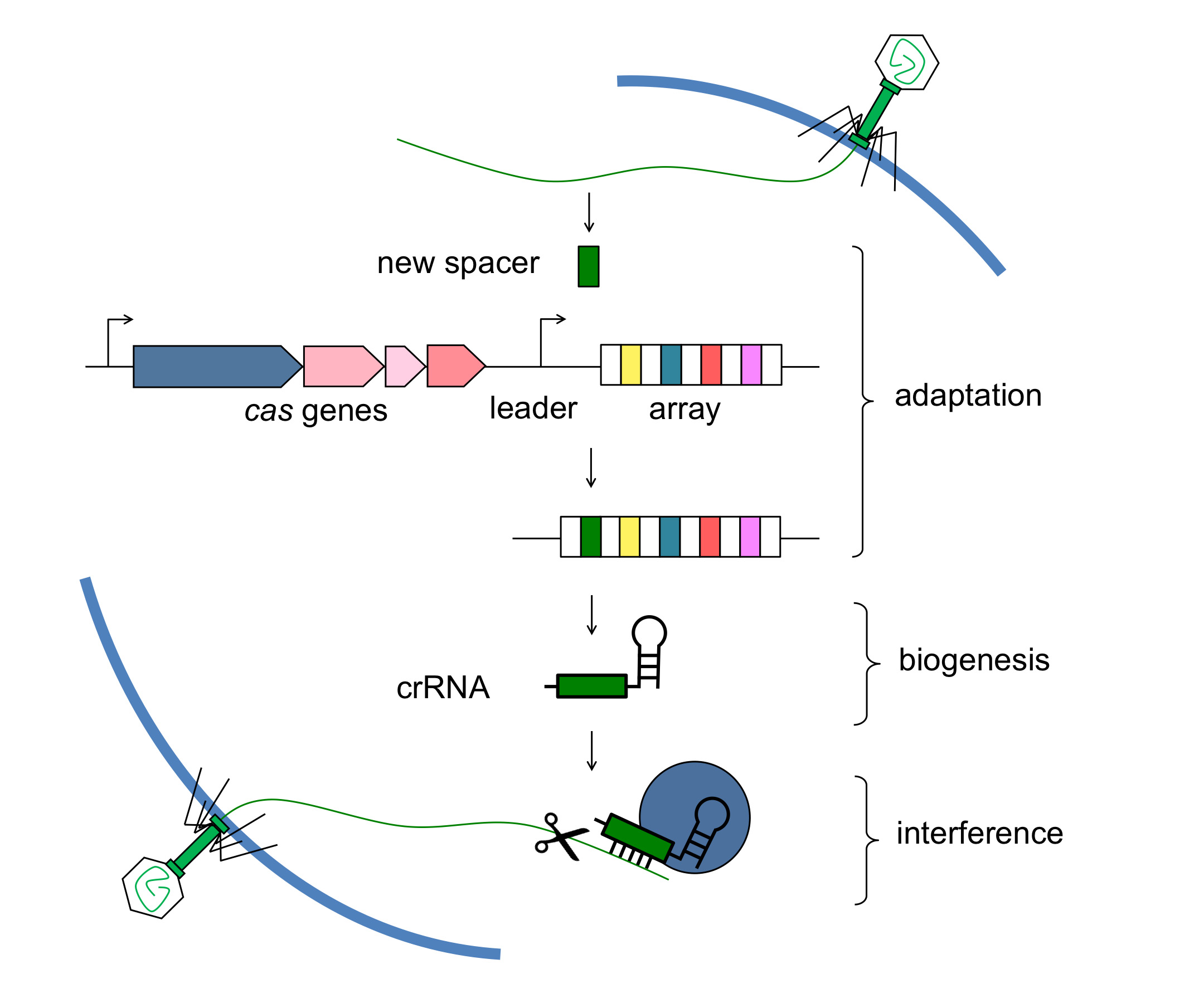
Sequence-directed genetic interference pathways control gene expression and preserve genome integrity in all kingdoms of life. In many bacteria and most archaea, clustered, regularly interspaced, short palindromic repeats (CRISPRs) specify a recently discovered genetic interference pathway that protects cells from viruses (phages) and conjugative plasmids. Within CRISPR sequences, the repeats are separated by short spacer sequences that match phage or plasmid genomes and specify the targets of interference. Spacer sequences are transcribed into CRISPR RNAs (crRNAs) — small RNAs that, through base-pairing interactions with the target sequence, guide Cas nucleases to the invasive nucleic acid. Upon infection, CRISPR arrays can acquire new repeat-spacer units that match the infecting phage or plasmid. Therefore CRISPR-Cas systems provide adaptive and inheritable immunity to the bacterial cell. The spacer content of CRISPR arrays reflects the many different invaders encountered by the host and can be expanded rapidly in response to new ones. Accordingly, CRISPR loci constitute a “genetic memory” that ensures the rejection of new, returning and ever-present invading DNA molecules.
We use Staphylococcus epidermidis and Streptococcus pyogenes as model systems for studying CRISPR immunity. The clinical isolate S. epidermidis RP62a harbors a CRISPR spacer that matches the nickase gene (nes) present in nearly all staphylococcal conjugative plasmids carrying antibiotic resistance genes and prevents their spread. Using this system, Dr. Marraffini revealed that the CRISPR-Cas machinery targets DNA, rather than RNA, directly. Work in the Marraffini lab also demonstrated that the S. pyogenes crRNA-guided Cas9 DNA nuclease constitutes a formidable tool for genetic engineering.
The lab’s current research employs molecular genetic and biochemical approaches to analyze the genesis and function of CRISPR-Cas systems. We ultimately hope to answer fundamental questions about how CRISPR-Cas systems destroy their targets, how the genetic memory is generated, and how CRISPR-Cas immunity affects the evolution of bacteria and archaea, particularly of bacterial pathogens. By performing detailed analyses of the underlying molecular mechanisms of CRISPR immunity, we hope to facilitate the manipulation of this natural pathway for the development of new technologies.